IN SILICO ANALYSIS OF PHAG-LIKE PROTEIN IN RALSTONIA
EUTROPHA H16, POTENTIALLY INVOLVED IN
POLYHYDROXYALKANOATES SYNTHESIS
Melissa
Uribe Acosta1, Andrés Felipe Villa Restrepo 1
1
Grupo
Biotransformación,
Escuela de Microbiología, Universidad de Antioquia. Calle 67
n°. 53-108, laboratorio 5-226. Medellín, Colombia.
Correspondencia:
melissa.uribe1@udea.edu.co
ABSTRACT
Polyhydroxyalkanoates (PHA) are synthesised by
bacteria as carbon storage material. The protein PhaG directs
carbon from non-related carbon sources such as glycerol,
metabolised through fatty acid de novo synthesis (FAS)
pathway, with PHA synthesis. The gene that codifies for this
protein has not yet been found in the genome of Ralstonia
eutropha H16, a model organism. By bioinformatic
comparison to already known PhaG proteins, a PhaG-like protein
was found codified by gene H16_A0147 and presence of the gene
was preliminary confirmed by PCR. This is the first study that
shows the presence and characteristics of a PhaG-like protein
in R. eutropha H16 and represents the first step for
the identification of a connection between FAS and PHA
pathways in this model bacterium. Further gene deletion and
enzymatic activity studies are necessary to confirm this
potential relationship, which could improve industrial PHA
production and utilisation of agro-industrial residues such as
glycerol.
Keywords: Polyhydroxyalkanoates, Ralstonia eutropha
H16, non-related carbon sources, protein function prediction.
Recibido:
30 de Septiembre de 2019. Aceptado: 16 de Mayo de 2019
Received: September 30, 2019. Accepted: May 16, 2019
ANÁLISIS IN SILICO DE UNA PROTEÍNA
SIMILAR A PHAG EN RALSTONIA EUTROPHA H16
POTENCIALMENTE INVOLUCRADA EN LA SÍNTESIS DE
POLIHIDROXIALCANOATOS
RESUMEN
Los
polihidroxialcanoatos
(PHA) son sintetizados por las bacterias como material de
reserva de carbono. La proteína PhaG dirige el carbono proveniente
de fuentes de carbono no relacionadas como el glicerol, que
son metabolizados a través de la síntesis de ácidos grasos de novo (FAS), hacia la síntesis de PHA. El
gen que codifica esta proteína no ha sido aún encontrado en
el genoma de Ralstonia eutropha H16, un organismo
modelo. A
través de la comparación con proteínas PhaG ya conocidas,
una proteína similar a PhaG, fue encontrada siendo
codificada por el gen H16_A0147 y la presencia del gen
confirmada preliminarmente utilizando PCR. Este es el primer
estudio que muestra la presencia y características de una
proteína similar a PhaG en R.
eutropha H16 y representa el primer paso en la
identificación de una conexión entre las rutas metabólicas
FAS y de PHA en esta bacteria modelo. Estudios de bloqueo de
genes y actividad enzimática son necesarios para confirmar
esta relación potencial que podría mejorar la producción
industrial de PHA y la utilización de residuos
agroindustriales como el glicerol.
Palabras
clave:Polihidroxialcanoatos,
Ralstonia eutropha H16,
fuentes de carbono no relacionadas, predicción de función de
proteínas.
Cómo citar este artículo: M. Uribe,
A. Villa. “In silico analysis of phag-like protein in
ralstonia eutropha H16, potentially involved in
polyhydroxyalkanoates synthesis”, Revista Politécnica, vol.
15, no.29 pp.55-64, 2019. DOI: 10.33571/rpolitec.v15n29a5
Polyhydroxyalkanoates (PHA) are an environmentally friendly
alternative to the excessively used petrochemical plastics since
they are biodegradable and can be used for similar purposes such
as manufacturing of packaging materials or biomedical devices. PHA
are polyesters synthesised by bacteria as carbon storage compounds
when levels of oxygen, nitrogen or phosphorus are low. PHA can be
classified according to the amount of carbon atoms in the
hydroxyacyl-CoA monomers as short-chain-length PHA (3 to 5) and
medium-chain-length PHA (mlc-PHA) (6 to 14). The composition of
the monomers depends on the microorganism and the carbon source
used, since the latter can be transformed into hydroxyacyl-CoA
precursors by different metabolic routes [1, 2, 3].
Fatty acid de novo synthesis (FAS)
pathway is particularly interesting because these carbon
sources are generally present in inexpensive organic residues,
such as glycerol, which is a by-product of biodiesel
production [5, 6, 7]. Additionally, the polymer produced
through this pathway, mcl-PHA, can be used as a biodegradable
alternative for elastomer and rubber in cosmetics, paint
formulations and medical devices [8]. PhaG is the enzyme that
allows this connection between FAS pathway and PHA synthesis
by transforming the intermediate 3-hydroxyacyl-ACP into
3-hydroxyacyl-CoA [5].
PhaG was characterised for the first time by Rehm
et al. (1998) [5] in Pseudomonas putida KT2448
as a hydroxyacyl-CoA-ACP-transferase, however, Wang et al.
(2012) [7] suggested that the PhaG protein function is rather
a thioesterase. Bacteria missing this PhaG protein accumulated
85 % polymer with octanoate as substrate but only 3 % when
gluconate, which is metabolised through FAS pathway, was
provided as carbon source [5], indicating its importance for
PHA synthesis from gluconate. Furthermore, PHA production from
simple carbon sources was re-established in P. oleovorans
ATCC 29347 and P. fragi [9] and augmented up to 40 %
in P. aeruginosa PAO1 [5], only by the insertion of
the genes phaC + phaG or only phaG,
respectively. PhaG protein has only been experimentally
characterised and reported in Pseudomonas species, Burkholderia
caryophylli and Aeromonas hydrophila [10], out
of the 75 bacterial genera that have been reported as PHA
producers [2].
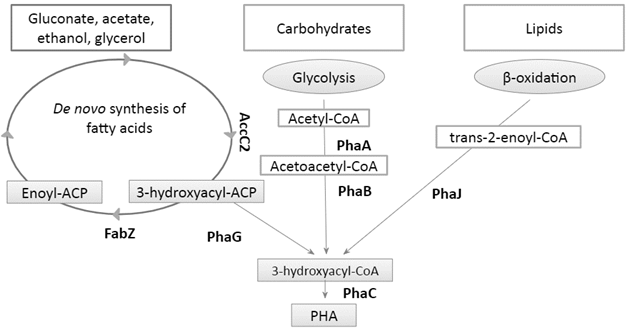
Fig. 1. Connection of central metabolic pathways with PHA
metabolism.
There has not been evidence that R. eutropha H16, the
most well-studied bacterium regarding PHA metabolism and model
for large-scale production, possesses a phaG
homologue. However, R. eutropha H16 has also shown its
ability to utilise alternative carbon sources such as
gluconate, glycerol or acetate for PHA synthesis [11].
Peplinski et al. (2010) [12] reported a relationship
between (FAS) and PHA synthesis, based on the upregulation of
genes involved in FAS pathway, such as accC2 and fabG,
when R. eutropha H16 grew on sodium gluconate as
carbon source and produced PHA. Additionally, other studies
have shown that deletion of up to 9 phaA homologues
does not suppress PHA production from sodium gluconate in this
bacterium [13, 14]. These evidences suggest that R.
eutropha H16 may possess proteins which are able to
perform the same function as PhaG.
Proteins with < 50 % similarity can be
compared on the basis of conserved motifs which can reveal
protein function even when this is not globally similar to any
known protein [15]. Protein function prediction is a major
issue in biology since the protein databases grow
exponentially with the crescent availability of fast and
inexpensive sequencing techniques while experimental protein
characterisation techniques are still time-consuming and
expensive. Due to in silico analysis, novel and useful
proteins can be targeted that will allow the development of
new PHA production strategies. The goals of this research
were: to identify genes in R. eutropha H16 codifying
proteins with high similarity to PhaG, to analyse and compare
those proteins in silico to all PhaG proteins that are
already experimentally characterised and to preliminarily
confirm the presence of the genes codifying these in
silico predicted proteins.
2.
METHODOLOGY
In silico
analysis and PhaG homologues characterisation
Using PhaG protein sequence from P. putida
KT2440 (Model PhaG protein [10]), accession number AAC34749.1,
a standard and specialised protein BLAST (BLASTP) was carried
on BacMap protein database, on both R. eutropha H16 chromosomes
(Matrix: BLOSUM62, Mask: low complexity, Program: blastp,
Database: Protein). Only those proteins with similarity >
40 % and e-value ≤ 1e-04 were selected. After a literature
search and selection for already characterised PhaG proteins
[10, 16], 9 PhaG homologues were analysed using the tool MOTIF
finder from the GenomeNet Database Resources of the Kyoto
University Bioinformatics Center. PhaG proteins for PROSITE
patterns, NCBI Conserved Domains (CDD) and Protein families
(Pfam) were aligned using the UniProt alignment tool
(www.uniprot.org/align) and conserved aminoacids related to
known catalytic aminoacid residues in thioesterases were
located manually. Using Membrane protein IdeNtificatioN
withOUt explicit use of hydropathy profiles and alignments
(MINNOU server), an image from the secondary structure of the
proteins was generated.
Culture conditions
R. eutropha H16 was maintained on tryptic soy agar (TSA)
plates. Individual colonies were inoculated in tryptic soy
broth (TSB) and incubated at 30 °C for 12 h. 2 mL of cultures
with OD600: 0,5, were centrifuged, supernatant was discarded
and cell pellet was washed twice in NaCl 0,9 % solution before
DNA extraction.
DNA extraction and R. eutropha H16 phaG-like
gene amplification
R. eutropha H16 DNA was used as a template for the
development and optimisation of phaG-like genes
amplification. DNA was obtained by using DNeasy Blood and
Tissue Kit (Qiagen) according to the manufacturer's protocol
for Gram negative bacteria.
Primers targeting phaG-like gene H16_A0147
were designed and the
desired sequence was sent to the company Eurofins scientific
to be synthesised. A Polymerase chain reaction (PCR) with
different annealing temperatures was carried out to determine
the annealing temperature. PCR
amplifications were performed in a 20 μL reaction mixture
containing 1X Taq buffer; 1.5 mM MgCl2, 0.2 mM of
dNTPs, 1 μM of each primer, 0.03 U/ μL of Taq DNA polymerase
and 1 μL of genomic DNA (25–30 ng). PCR conditions were:
initial denaturation at 95 °C for 30 s; denaturation at 95
°C for 20 s; annealing at 53-60 °C for 45 s; extension at 72
°C for 60 s (30 cycles) and final extension at 72 °C for 120
s. Four negative controls with no DNA were included at
annealing temperatures of 54, 56, 58 and 60. The resulting
PCR products were visualised in 1 % agarose gel and stained
with EZ-Vision® in gel solution.
3.
RESULTS
In silico
analysis and PhaG homologues characterisation.
Two proteins, one codified by a gene from
chromosome 1 and one codified by a gene from chromosome 2,
were found with % sequence identity >20, % positive
substitutions >40 and e-value <=1e-04 when compared to
PhaG sequence: (1) gene and locus tag H16_A014, with protein
accession number in NCBI CAJ91299.1 presented 21 %
identity and 41 % similarity with an e-value of 6x10e-4, is
located in chromosome 1 and has a gene size of 843 bp.
(2) gene mhpC with locus tag H16_B1070 and
protein accession number in NCBI CAJ95861.1 presented 22 %
identity and 40 % similarity with an e-value of 6x10e4, is
located in chromosome 2 and has a gene size of 795 bp.
According to Russell et al. (1997) [17]
proteins that are able to perform the same function, either a
remote homologue or analogue, can show less than 50 % of
sequence similarity due to shared active sites or conserved
domains; the formal definition of remote homology is protein
sequences that share an identity percentage of less than 25 %
[18]. mhpC gene is already annotated in R.
eutropha H16 genome as aminoacrylate hydrolase and
located in chromosome 2 while all PHA-related genes are
located in chromosome 1 in R. eutropha H16 [11]; for
those reasons, it was not included in further analysis as a
potential PhaG analogue or remote homologue.
Already characterised PhaG proteins as well as
H16_A0147 were individually tested for conserved domains and
protein families. All PhaG homologues and H16_A0147 were found
to have an α/β hydrolase1 domain as the principal and only
domain in CDD. No
matches were found for any of the proteins when using the
PROSITE database. All PhaG homologues, as well as H16_A0147,
exhibit significant similarity to Pfam Abhydrolase_6 and
Hydrolase_4 (Table 1). No PhaG homologue showed significant
similarity to a transferase Pfam or CDD. Interestingly, PhaG
from Pseudomonas sp. USM 4-55 as well as H16_A0147
showed significant similarity to a PHA depolymerase domain.
Table 1. E-values from PhaG homologues and R.
eutropha H16_RS00705 compared to Pfam database.
|
Strains
|
Abhydrolase_1
|
Abhydrolase_6
|
Hydrolase_4
|
Accession number
|
|
R. eutropha H16 (H16_A0147)
|
1.30E-28
|
3.80E-17
|
6.70E-14
|
CAJ91299.1
|
|
P. putida KT2447
|
5.40E-14
|
2.40E-06
|
9.90E-09
|
AAC34749.1
|
|
Pseudomonas sp. USM 4-55
|
2.00E-10
|
5.10E-11
|
4.20E-10
|
ACA03779.1
|
|
P. aeruginosa
|
2.00E-10
|
3.20E-08
|
1.50E-07
|
AAF61903.1
|
|
P. stutzeri strain 1317
|
5.40E-14
|
2.40E-06
|
9.90E-09
|
AAM64206.2
|
|
P. nitroreducens strain 0802
|
8.70E-14
|
6.50E-06
|
3.60E-08
|
AAK71349.1
|
|
P. mendocina strain LZ
|
1.30E-11
|
8.60E-06
|
1.60E-06
|
AAQ16175.1
|
|
P. oleovorans
|
4.50E-13
|
2.30E-06
|
1.50E-08
|
AAF89663.1
|
|
Burkholderia caryophylli
|
5.20E-10
|
>1E-4
|
3.50E-07
|
AAK71350.1
|
|
Pseudomonas sp. 61-3
|
7.10E-09
|
>1E-4
|
1.50E-08
|
BAB32432.1
|
|
P. fluorescens strain BM07
|
6.90E-07
|
>1E-4
|
2.00E-07
|
ACA60824.1
|
Confirmation of H16_A0147 gene presence
A PCR product with around 800 bp was obtained
using the designed primers on DNA extracted from R.
eutropha H16, in accordance with the in silico
obtained information about H16_A0147 sequence (Table 2).
Table 2. Amplification of a phaG-like
gene from R. eutropha H16 genome on gradient
temperature PCR.
|
Forward primer
|
5’-CACGCCACCAGCCGAAA-3’
|
|
Reverse primer
|
5´-GATTGGATCCTCACGGAACGTCG -3
|
|
Annealing temperature used
|
53
|
54
|
55
|
56
|
57
|
58
|
59
|
60
|
|
Presence/absence of PCR product (+/-)
|
-
|
-
|
-
|
-
|
-
|
+
|
+
|
+
|
|
Approximate PCR product length obtained
(bp)-
|
|
-
|
-
|
-
|
-
|
700-1000
|
700-1000
|
700-1000
|
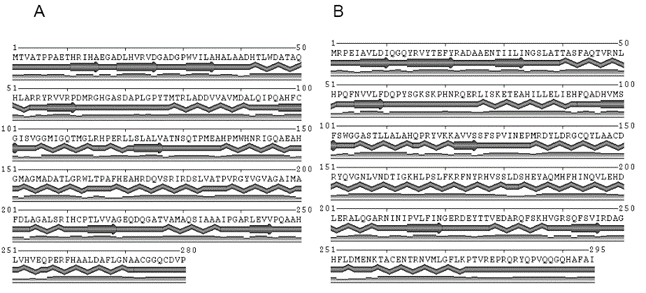
Fig. 2. Predicted secondary structure image generated with
MINNOU Server [19], β-sheets are represented
as arrows and α-helices as waves. The third line indicates
the confidence level of the predicted structure for
that particular position.
4. DISCUSSION
In silico
analysis and PhaG homologues characterisation.
The comparison of the Pfam analysis among the
PhaG homologues and H16_A0147 indicates they belong to the
same protein family, α/β hydrolase fold family, in spite of the low sequence similarity. This
fact further suggests they might share catalytic active sites.
Regarding the H16_A0147 gene, a thioesterase domain [20] was
found in its sequence but no domain indicating a transferase
function.
PhaG had been originally classified as an
(R)-3-hydroxyacyl- ACP-CoA transferase based on the ability of
a partially purified extract to convert 3-hydroxydecanoyl-CoA
into 3-hydroxydecanoyl-ACP when ACP was present [5]. However,
overexpression of phaG in E. coli resulted in
the extracellular accumulation of 3-hydroxydecanoic acid [7]
and low PHA production (0,9 mg/L) compared to E. coli
harbouring both phaG and PP0763 (25 mg/L) a predicted
medium-chain-fatty-acid CoA ligase [21]. Those results suggest
that PhaG has rather thioesterase activity than
transferase activity and separates the ACP moiety from the
3-hydroxyacyl before another enzyme, probably a ligase,
catalyses the merging of the CoA moiety to form
3-hydroxyacyl-CoA. Our results are according to those from Wang et
al. (2012) [21] and, since no transferase conserved
domain was found, support the possibility that PhaG-like
proteins in R. eutropha H16 perform a thioesterase
function instead of a transferase function. Further studies
are necessary in order to characterise the enzyme activity of
this protein.
α/β hydrolase fold family is one of the largest
groups of structurally related proteins. Thioesterases are
included together with other hydrolytic enzymes such as
acetylcholinesterases, carboxylesterases, dienelactones
hydrolases, lipases, cutinases, serine carboxypeptidases,
proline iminopeptidases, proline oligopeptidases and epoxide
hydrolases, and also enzymes that require HCN, H2O2
or O2 instead of H2O such as haloalkane
dehalogenases, haloperoxidases and hydroxynitrile lyases [22]
but the family also includes non-catalytic enzymes [23].
Thioesterases can hydrolyse the ester bond between a carbonyl
group and a sulphur atom, the substrates include both CoA and
ACP moieties, and are classified in 23 families with low
sequence homology but similar tertiary structure [20].
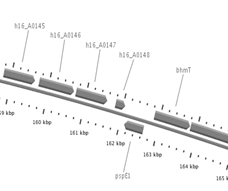
The genome context was also analysed for
H16_A0147 gene and compared with the location of phaG
from P. putida KT 2447. H16_A0147 gene is surrounded
upstream by other genes codifying hypothetical proteins, and
downstream by gene pspE1 codifying a rhodanese-related
sulphurtransferase and by bhmT codifying a
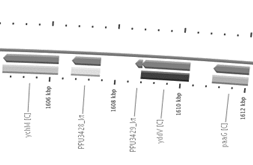
methyltransferase (Figure 3). Unlike H16_A0147, phaG gene
is located in the lagging strand and is surrounded upstream by
a gene codifying a hypothetical protein and by gene yddV
codifying a diguanylate cyclase. Downstream it is surrounded
by gene ychM codifying a sulphate transporter and by a
gene codifying a hypothetical protein (Figure 5). No
similarities were found regarding the genome context of the
two genes.
Confirmation of PhaG-like gene presence
Using the primers designed the in silico
sequence to target specifically the gene H16_A0147, a PCR
product was obtained with the expected length, at annealing
temperatures of 58, 59 and 60 °C, indicating lower
temperatures are not optimal for primer alignment to the DNA
targeted region. It is necessary to perform DNA sequencing on
this PCR product in future studies to confirm that the
sequence belongs to this particular gene. This confirmation
could imply that this microorganism has an important element
for the metabolic pathway implicated in the connection of FAS
with PHA synthesis.
FAS pathway is necessary to provide fatty acids
to the cell, required for phospholipid synthesis and,
therefore, membrane formation for cell growth and division -
functions that are highly active during exponential phase and
decline during stationary phase when PHA production begins
[12, 25, 26]. However, among these studies, Peplinski et
al. (2010) [12] and Shimizu et al. (2013) [26]
reported genes accC2 and fabG involved in FAS
pathway were upregulated during PHA production in P.
putida KT2447. These genes are expressed during the
initial steps of FAS, and could allow accumulation of
3-hydroxyacyl-ACP, which is the substrate for PhaG. According
to that, Wang et al. (2012) [21] demonstrated that phaG,
phaC1 and phaC2 genes expression showed n-fold
induction of 220, 2.6 and 4.3 respectively, during PHA
production phase in the same bacteria. We suggest this could
be happening in R. eutropha H16 metabolism, and could
be further studied by gene expression analysis targeting
H16_A0147, accC2 and fabG.
A
B
Fig. 3. Localisation and surrounding genes in
their respective strains for (A) H16_A0147 in R. eutropha
H16 genome and (B) phaG (PPU3428_kt) in P.
putida KT2447 genome (Source: Bacmap [24]).
This is the first study that shows the presence
and the characteristics of a putative phaG-like gene
in R. eutropha H16 and represents the first step for
the identification of a possible connection between FAS
pathway and PHA synthesis in this novel bacterium. Further
studies are necessary to confirm the potential relationship of
the gene product of H16_A0147 gene with the PHA metabolism.
Characterising a PhaG-like protein in R. eutropha H16
could allow the use of residues from biodiesel industry [6,
7], or phenylacetic acid, which is a contaminating compound
[27], for PHA synthesis in the future.
5. CONCLUSIONS
H16_A0147 was identified as a potential gene
codifying for a PhaG-like protein with Thioesterase activity.
Further studies are necessary to confirm this metabolic link.
If this link is confirmed, expression of H16_A0147 may be
manipulated in R. eutropha H16 for the production of
PHA using waste products such as glycerol or contaminants like
phenylacetic acid as carbon sources.
6.
ACKNOWLEDGEMENTS
Carolina Ramírez, Luisa Múnera, Dr. Nancy Pino,
the School of Microbiology, laboratory GDCON, all from
University of Antioquia, and four anonymous evaluators
contributed to this study and are gratefully acknowledged.
7. REFERENCES
[1] Aldor I.S. and Keasling, J.D. (2003). Process Design for
Microbial Plastic Factories: Metabolic Engineering of
Polyhydroxyalkanoates. Curr. Opin. Biotechnol. 14 475–483.
DOI: 10.1016/j.copbio.2003.09.002
[2] Reddy, C.S.K. Ghai, R., Rashmi. and Kalia,
V.C. (2003). Polyhydroxyalkanoates: An Overview. Biores
Tech 87, 137–146. DOI:
10.1016/S0960-8524(02)00212-2
[3]
Urtivia,
V., Villegas, P., González, M. and Seeger, M. (2014). Bacterial Production of the Biodegradable
Plastics Polyhydroxyalkanoates. Intern J Biol Macromol.
70: 208–213. DOI: 10.1016/j.ijbiomac.2014.06.001
[4] Lau, N., Foong, C.P., Kurihara, Y., Sudesh,
K. and Matsui, M. (2014). RNA-Seq Analysis Provides Insights
for Understanding Photoautotrophic Polyhydroxyalkanoate
Production in Recombinant Synechocystis Sp. PLOS
one. Vol 9(1). DOI: 10.1371/journal.pone.0086368
[5] Rehm, B.H.A., Kruger, N. and Steinbuchel, A.
(1998). A New Metabolic Link between Fatty Acid de
novo Synthesis and Polyhydroxyalkanoic Acid Synthesis:
The phaG Gene from Pseudomonas putida
KT2440 Encodes a 3-hydroxyacyl- acyl carrier Protein-coenzyme
a Transferase. J. Biol. Chem. 273:24044-24051. DOI: 10.1074/jbc.273.37.24044
[6] Nomura, C.T., Tanaka, T., Eguen, T.E., Appah,
A.S., Matsumoto, K., Taguchi, S., Ortiz, L. and Doi, Y.
(2008). FabG Mediates Polyhydroxyalkanoate Production from
Both Related and Nonrelated Carbon Sources in Recombinant Escherichia
coli LS5218. Biotechnol. 24: 42-351. DOI: 10.1021/bp070303y
[7] Wang, Q., Zhuang, Q., Liang, Q. and QI, Q.
(2013). Polyhydroxyalkanoic Acids from
Structurally-unrelated Carbon Sources in Escherichia coli.
Appl Microbiol Biotechnol. Vol 97:3301–3307.
DOI: 10.1007/s00253-013-4809-x
[8] Leong, Y.K., Show P.L., Ooi, C.W., Ling, T.C.
and Lan, J.C. (2014). Current Trends in Polyhydroxyalkanoates
(PHAs) Biosynthesis: Insights from the Recombinant Escherichia
coli. J Biotech 180, 52-65. DOI: 10.1016/j.jbiotec.2014.03.020
[9] Fiedler, S., Steinbuchel, A. and Rehm, B.H.
(2000). PhaG-Mediated Synthesis of
Poly(3-Hydroxyalkanoates) Consisting of Medium-Chain- Length
Constituents from Nonrelated Carbon Sources in Recombinant Pseudomonas
fragi. Appl. Environ. Microbiol.
66(5):2117-2124. DOI: 10.1128/aem.66.5.2117-2124.2000
[10] Röttig, A., and Steinbüchel, A. (2013).
Acyltransferases in Bacteria. Microbiol Mol Bio Reviews :
MMBR, 77(2), 277–321. DOI: 10.1128/MMBR.00010-13
[11] Pohlmann, A., Fricke, W. F., Reinecke, F.,
Kusian, B., Liesegang, H., Cramm, R., Eitinger, T., Ewering,
C., Potter, M., Schwartz, E., Strittmatter, A., Voss, I.,
Gottschalk, G., Steinbuchel, A., Friedrich, B. and Bowien, B.
(2006). Hydrogen-based Biotechnology: Genome Sequence of the
Bioplastic-producing ‘‘Knallgas’’ Bacterium Ralstonia
eutropha H16. Nat Biotechnol 24, 1257–1262. DOI: 10.1038/nbt1244
[12] Peplinski, K., Ehrenreich, A., Doring, C.,
Bomeke, M., Reinecke, F., Hutmacher, C. and Steinbuchel, A.
(2010). Genome-wide Transcriptome Analyses of the
‘Knallgas’ Bacterium Ralstonia eutropha H16 with
Regard to Polyhydroxyalkanoate Metabolism. Microbiology, 156: 2136–2152. DOI: 10.1099/mic.0.038380-0
[13] Lindenkamp, N., Peplinski, K., Volodina, E.,
Ehrenreich, A. and Steinbuchel, A. (2010). Impact of Multiple Beta-Ketothiolase Deletion
Mutations in Ralstonia eutropha H16 on the Composition
of 3-Mercaptopropionic Acid-Containing Copolymers. Appl. Environ. Microbiol. Vol.
76(16): 5373–5382. DOI: 10.1128/AEM.01058-10
[14] Lindenkamp, N., Volodina, E. and Steinbuchel,
A. (2012). Genetically Modified Strains of Ralstonia
eutropha H16 with Beta-Ketothiolase Gene Deletions for
Production of Copolyesters with Defined 3-Hydroxyvaleric Acid
Contents. App. Environ. Microbio. 78(15):5375-83.
DOI: 10.1128/AEM.00824-12
[15] Nevill-Manning, C., Wu, T. and Brutlag, D.
(1998). Highly Specific Protein Sequence Motifs for Genome
Analysis. Proc. Natl Acad. Sci. 95:5865–5871. DOI:
10.1073/pnas.95.11.5865
[16] Arsad H. (2009). Cloning and
Characterisation of (R)-3-hydroxyacyl-acyl Carrier
Proteincoenzyme A Transferase Gene (phaG) from Pseudomonas sp.
USM 4-55. Trop. Life. Sci. Res. 20(2):1-14.
[17] Russell, B., Saqi, M.A., Sayle, R.A., Bates,
P.A. and Sternberg, M.J.E. (1997). Recognition of Analogous
and Homologous Protein Folds: Analysis of Sequence and
Structure Conservation. J. Mol. Biol. 269:423-439.
DOI: 10.1006/jmbi.1997.1019
[18] Bedoya, O. and Tischer, I. (2015). Reducing Dimensionality in Remote Homology
Detection Using Predicted Contact Maps. Comput. Biol. Med.
59:64-72. DOI: 10.1016/j.compbiomed.2015.01.020
[19] Cao, B., Porollo, A., Adamczak, R., Jarrell,
M. and Meller, J. (2006). Enhanced Recognition of Protein
Transmembrane Domains with Prediction-based Structural
Profiles. Bioinformatics. 22:303-9. DOI:
10.1093/bioinformatics/bti784
[20] Cantu, D.C., Chen, Y. and Reilly, P.J.
(2010). Thioesterases: A New Perspective Based on their
Primary and Tertiary Structures. Protein Sci.
19:1281–1295. DOI: 10.1002/pro.417
[21] Wang, Q., Tappel, R.C., Zhu, C. and Nomura, C.T.
(2012). Development of a New Strategy for Production of
Medium-chain-length Polyhydroxyalkanoates by Recombinant
Escherichia coli via Inexpensive Non-fatty Acid Feedstocks. Appl
Environ Microbiol. 78(2):519-27. DOI: 10.1128/AEM.07020-11
[22] Bugg, T.D. (2004). Diverse Catalytic
Activities in the Alphabeta-hydrolase Family of Enzymes:
Activation of H2O, HCN, H2O2, and O2. Bioorg Chem.
32(5):367-375. DOI: 10.1016/j.bioorg.2004.05.005
[23] Nardini, M. and Dijkstra, B.W. (1999). α/β
Hydrolase Fold Enzymes: The Family Keeps Growing. Curr Opin
Struct Biol. 9(6):732-737.
[24] Stothard, P., Van Domselaar, G.,
Shrivastava, S., Guo, A., O'Neill, B., Cruz, J., Ellison, M.
and Wishart, D.S. (2005). BacMap: An Interactive Picture Atlas
of Annotated Bacterial Genomes. Nucleic Acids Res
33:D317-D320.
[25] Brigham, C.J., Speth, D.R., Rha, C. and
Sinskeya, A.J. (2012). Whole-Genome Microarray and Gene
Deletion Studies Reveal Regulation of the Polyhydroxyalkanoate
Production Cycle by the Stringent Response in Ralstonia
eutropha H16. App Environ Microbiol. 78;8033–8044. DOI:
10.1128/AEM.01693-12
[26] Shimizu, R., Chou, K., Orita, I., Suzuki,
Y., Nakamura, S. and Fukui, T. (2013). Detection of
Phase-dependent Transcriptomic Changes and Rubisco-mediated
CO2 Fixation into Poly (3-hydroxybutyrate) under Heterotrophic
Condition in Ralstonia eutropha H16 Based on RNA-seq and Gene
Deletion Analyses. BioMed Cen Microbiol. 13:169. DOI:
10.1186/1471-2180-13-169
[27] Tobin, K.M., O'Leary, N.D., Dobson, A.D. and
O'Connor, K.E. (2007). Effect of Heterologous Expression of
phaG [(R)-3-hydroxyacyl-ACP-CoA transferase] on
Polyhydroxyalkanoate Accumulation from the Aromatic
Hydrocarbon Phenylacetic Acid in Pseudomonas species. FEMS
Microbiol Lett. 268(1):9-15. DOI:
10.1111/j.1574-6968.2006.00607.x



